In the era of big data and artificial intelligence, the ability to store, manage, and retrieve vast amounts of data efficiently is more crucial than ever. Vector databases have emerged as a powerful tool in handling data in the form of vectors, facilitating efficient search and retrieval operations essential for AI and machine learning applications. Central to the performance of these databases are the indexing techniques they employ and the computational efficiency they achieve, often leveraging advanced hardware like GPUs and TPUs. This post delves into how various indexing techniques relate to vector databases and the role of efficient computation in optimizing their performance.
Table of contents
Open Table of contents
Vector Databases
Vector databases are specialized systems designed to store and manage data represented as high-dimensional vectors. These vectors often originate from feature extractions in machine learning models, such as embeddings from natural language processing or image recognition tasks. Efficiently searching and retrieving similar vectors is vital for applications like recommendation systems, semantic search, and anomaly detection.
A vector is a one-dimensional array of numbers that represents a sequence of values. It has a rank of n=1. Although often visualized as a row for convenience, vectors are usually considered as columns in mathematical notation.
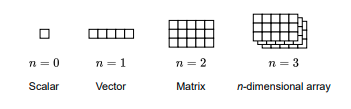
Embedding and indexing
Embedding is the process of turning raw data into vectors, which can then be indexed and searched over. Meanwhile, indexing is the process of creating and maintaining an index over vector embeddings, a data structure that allows for efficient search and information retrieval from a dataset of embeddings.
An embedding is a compact representation of raw data, such as an image or text, transformed into a vector comprising floating-point numbers. It’s a powerful way of representing data according to its underlying meaning. High-dimensional data is mapped into a lower-dimensional space (think of it as a form of “lossy compression”) that captures structural or semantic relationships within data, making it possible for embeddings to preserve important information while reducing the computational burden that comes with processing large datasets. It also helps uncover patterns and relationships in data that might not have been apparent in the original space.
Since an embedding model represents semantically similar things close together (the more similar the items, the closer the embeddings for those items are placed in the vector space), you can now have computers search for and recommend semantically similar things and cluster things by similarity with improved accuracy and efficiency.
Efficient computation in vector database searches
Searches in vector databases require intensive calculations with large volumes of data. Graphical Processing Units (GPUs) are fundamental in accelerating these processes, enabling faster and more efficient operations. As their usage in AI and data processing has increased, GPUs have been equipped with dedicated tensor cores, and specialized chips like Google’s Tensor Processing Units (TPUs) have been developed.
A GPU possesses several thousand parallel units and its own fast memory, which can be leveraged to accelerate vector similarity searches. However, the limiting factor is usually not the number of computing units but the read-write operations to memory. The slowest link is often between the CPU memory and the GPU memory; consequently, copying data across devices should be minimized. Moreover, the GPU’s internal structure involves multiple levels of cache memory, which are smaller but faster, so computations should be organized to avoid unnecessary data transfers between these caches.
In the context of vector database searches, this is achieved by organizing computations in batches of queries or data samples that can fit entirely in GPU memory and be processed in parallel. When performing similarity searches, both the query vectors and the database vectors (or relevant indexes) need to be moved to the cache memory near the computing units. Processing queries in batches allows for copying the data only once, instead of doing it for each query individually. In practice, a GPU processes a batch that fits in memory almost as quickly as it would process a single query, significantly improving throughput.
A standard GPU has a theoretical peak performance of 10¹³–10¹⁴ floating-point operations (FLOPs) per second, and its memory typically ranges from 8 to 80 gigabytes. The standard FP32 encoding of floating-point numbers uses 32 bits, but empirical results show that using 16-bit encoding, or even less for some operands, does not degrade performance. In vector databases, using lower-precision representations can reduce memory usage and increase computation speed, allowing for faster searches over larger datasets.
The importance of indexing techniques
Indexing techniques are methods used to organize and optimize the storage of vectors in a database, significantly impacting the speed and accuracy of search operations. The right indexing strategy can reduce computational costs, enhance performance, and enable scalability to handle large datasets.
The interplay between indexing techniques and computational efficiency
Selecting the appropriate indexing technique is crucial for optimizing both the performance of vector databases and the efficient use of computational resources. Techniques like IVF, DiskANN, and SPANN are tailored to handle large-scale data efficiently by optimizing storage and search operations. They reduce computational overhead and make better use of hardware capabilities.
On the other hand, computational efficiency is not just about faster hardware but also about smarter algorithms that can leverage this hardware effectively. Advanced indexing techniques reduce unnecessary computations, allowing GPUs and TPUs to process data in batches more effectively, thus enhancing overall system performance.
The relationship between the presented fundamentals and the indexing techniques can be understood by examining how search operations are managed and optimized in vector databases, as well as the importance of computational efficiency in executing these searches.
Flat indexing
This is the simplest technique for storing vectors without any modifications. Although it guarantees precise results, it is not efficient for large databases due to high computational costs. This contrasts with the idea of optimization and efficiency sought in AI applications, where handling large amounts of data is crucial.
The need for computational resources is high, which may limit this technique in implementations that require optimized performance, even with advanced technologies such as GPUs and TPUs.
- Playground
Stores all vectors as they are without any modifications. Searches are performed by comparing a query vector with every vector in the dataset.
import numpy as np
# Create a dataset of 1000 random 128-dimensional vectors
np.random.seed(42)
dataset = np.random.random((1000, 128))
# Query vector
query_vector = np.random.random(128)
# Compute Euclidean distances between the query vector and all dataset vectors
distances = np.linalg.norm(dataset - query_vector, axis=1)
# Find the index of the nearest neighbor
nearest_neighbor_idx = np.argmin(distances)
print(f"The nearest neighbor is at index {nearest_neighbor_idx} with a distance of {distances[nearest_neighbor_idx]:.4f}")
# Flat Indexing:
# Indexing Time: 0.000001 seconds
# Indexing Memory Usage: 65.58 MiB
# Query Time: 0.003910 seconds
# Query Memory Usage: 82.52 MiB
# The nearest neighbor is at index 8769 with a distance of 3.6533Locality-Sensitive Hashing (LSH)
LSH addresses the inefficiencies of flat indexing by hashing vectors into buckets based on similarity. This reduction in search space accelerates query responses but may introduce some inaccuracies due to potential hash collisions.
By decreasing the number of comparisons, LSH leverages hardware capabilities more effectively. Batch processing on GPUs can handle hashed groups efficiently, but the method still requires careful resource management to maintain performance.
- Playground
Hashes vectors into buckets such that similar vectors are more likely to be in the same bucket, reducing the search space.
import numpy as np
import time
from memory_profiler import memory_usage
from sklearn.random_projection import SparseRandomProjection
from collections import defaultdict
# Parameters
dataset_size = 10000
dimensionality = 128
num_hash_tables = 5
hash_size = 10
# Generate dataset
np.random.seed(42)
dataset = np.random.random((dataset_size, dimensionality))
# Measure indexing time and memory usage
start_time = time.time()
index_memory_before = memory_usage()[0]
# Generate random projections for hashing
projections = [SparseRandomProjection(n_components=hash_size) for _ in range(num_hash_tables)]
# Fit the projections
for proj in projections:
proj.fit(dataset)
hash_tables = [defaultdict(list) for _ in range(num_hash_tables)]
for idx, vector in enumerate(dataset):
for i, proj in enumerate(projections):
hash_key = tuple((vector @ proj.components_.T > 0).astype(int))
hash_tables[i][hash_key].append(idx)
indexing_time = time.time() - start_time
index_memory_after = memory_usage()[0]
index_memory = index_memory_after - index_memory_before
# Query vector
query_vector = np.random.random(dimensionality)
# Measure query time and memory usage
start_time = time.time()
query_memory_before = memory_usage()[0]
candidate_indices = set()
for i, proj in enumerate(projections):
hash_key = tuple((query_vector @ proj.components_.T > 0).astype(int))
candidates = hash_tables[i].get(hash_key, [])
candidate_indices.update(candidates)
candidates = dataset[list(candidate_indices)]
distances = np.linalg.norm(candidates - query_vector, axis=1)
nearest_idx_in_candidates = np.argmin(distances)
nearest_neighbor_idx = list(candidate_indices)[nearest_idx_in_candidates]
query_time = time.time() - start_time
query_memory_after = memory_usage()[0]
query_memory = query_memory_after - query_memory_before
print(f"LSH Indexing:")
print(f"Indexing Time: {indexing_time:.6f} seconds")
print(f"Indexing Memory Usage: {index_memory:.2f} MiB")
print(f"Query Time: {query_time:.6f} seconds")
print(f"Query Memory Usage: {query_memory:.2f} MiB")
print(f"The nearest neighbor is at index {nearest_neighbor_idx}")
# LSH Indexing:
# Indexing Time: 1.513633 seconds
# Indexing Memory Usage: -2.94 MiB
# Query Time: 0.113924 seconds
# Query Memory Usage: 9.50 MiB
# The nearest neighbor is at index 3912
Inverted File Indexing (IVF)
IVF organizes vectors into clusters using algorithms like K-means clustering. This hierarchical approach allows the system to search within relevant clusters rather than the entire dataset. Variants like IVF_PQ (Product Quantization) and IVF_SQ (Scalar Quantization) further compress vectors, enhancing search speed and reducing memory usage.
IVF benefits from parallel processing capabilities of GPUs and TPUs. Compressed vectors reduce memory bandwidth requirements, allowing for faster batch processing and more efficient utilization of computational resources.
- Playground
Clusters the dataset using methods like K-means, then searches within the relevant clusters.
import numpy as np
import time
from memory_profiler import memory_usage
from sklearn.cluster import KMeans
from collections import defaultdict
# Parameters
dataset_size = 10000
dimensionality = 128
num_clusters = 100
# Generate dataset
np.random.seed(42)
dataset = np.random.random((dataset_size, dimensionality))
# Measure indexing time and memory usage
start_time = time.time()
index_memory_before = memory_usage()[0]
kmeans = KMeans(n_clusters=num_clusters, random_state=42)
cluster_indices = kmeans.fit_predict(dataset)
inverted_index = defaultdict(list)
for idx, cluster_idx in enumerate(cluster_indices):
inverted_index[cluster_idx].append(idx)
indexing_time = time.time() - start_time
index_memory_after = memory_usage()[0]
index_memory = index_memory_after - index_memory_before
# Query vector
query_vector = np.random.random(dimensionality)
# Measure query time and memory usage
start_time = time.time()
query_memory_before = memory_usage()[0]
closest_cluster_idx = kmeans.predict([query_vector])[0]
candidate_indices = inverted_index[closest_cluster_idx]
candidates = dataset[candidate_indices]
distances = np.linalg.norm(candidates - query_vector, axis=1)
nearest_idx_in_candidates = np.argmin(distances)
nearest_neighbor_idx = candidate_indices[nearest_idx_in_candidates]
query_time = time.time() - start_time
query_memory_after = memory_usage()[0]
query_memory = query_memory_after - query_memory_before
print(f"IVF Indexing:")
print(f"Indexing Time: {indexing_time:.6f} seconds")
print(f"Indexing Memory Usage: {index_memory:.2f} MiB")
print(f"Query Time: {query_time:.6f} seconds")
print(f"Query Memory Usage: {query_memory:.2f} MiB")
print(f"The nearest neighbor is at index {nearest_neighbor_idx} within cluster {closest_cluster_idx}")
# IVF Indexing:
# Indexing Time: 0.670112 seconds
# Indexing Memory Usage: 35.91 MiB
# Query Time: 0.105518 seconds
# Query Memory Usage: 0.02 MiB
# The nearest neighbor is at index 5125 within cluster 64
Disk-Based Approximate Nearest Neighbor (DiskANN)
DiskANN is designed for extremely large datasets that exceed main memory capacities. It uses a graph-based approach stored on SSDs to minimize disk reads during searches. By constructing a graph with a small search diameter, it ensures scalability without compromising too much on performance.
DiskANN maximizes the use of available hardware by balancing in-memory computations with disk I/O operations. Efficient algorithms reduce the dependence on disk speed, and when combined with GPUs, it can handle large-scale searches more effectively.
- Playground
Designed for large datasets that don’t fit in memory, using SSDs to store and search vectors efficiently.
NOTE: DiskANN is a specialized system that primarily exists as a C++ library, and there isn’t a direct Python implementation available for demonstration purposes. However, we can illustrate the concept by simulating disk-based storage using memory-mapped files.
import numpy as np
import time
from memory_profiler import memory_usage
import faiss
# Parameters
num_vectors = 100000 # Reduced for practical execution
dimensionality = 128
chunk_size = 10000
# Generate and save dataset
np.random.seed(42)
dataset = np.float32(np.random.random((num_vectors, dimensionality)))
np.save('dataset.npy', dataset)
# Measure indexing time and memory usage
start_time = time.time()
index_memory_before = memory_usage()[0]
# Memory-map the dataset
mapped_dataset = np.load('dataset.npy', mmap_mode='r')
index = faiss.IndexFlatL2(dimensionality)
# Indexing in chunks to simulate disk-based indexing
for i in range(0, num_vectors, chunk_size):
data_chunk = np.array(mapped_dataset[i:i+chunk_size])
index.add(data_chunk)
indexing_time = time.time() - start_time
index_memory_after = memory_usage()[0]
index_memory = index_memory_after - index_memory_before
# Query vector
query_vector = np.float32(np.random.random((1, dimensionality)))
# Measure query time and memory usage
start_time = time.time()
query_memory_before = memory_usage()[0]
D, I = index.search(query_vector, k=1)
query_time = time.time() - start_time
query_memory_after = memory_usage()[0]
query_memory = query_memory_after - query_memory_before
print(f"Disk-Based ANN Simulation:")
print(f"Indexing Time: {indexing_time:.6f} seconds")
print(f"Indexing Memory Usage: {index_memory:.2f} MiB")
print(f"Query Time: {query_time:.6f} seconds")
print(f"Query Memory Usage: {query_memory:.2f} MiB")
print(f"The nearest neighbor is at index {I[0][0]} with a distance of {D[0][0]:.4f}")
# Disk-Based ANN Simulation:
# Indexing Time: 0.176873 seconds
# Indexing Memory Usage: 130.80 MiB
# Query Time: 0.115020 seconds
# Query Memory Usage: 0.50 MiB
# The nearest neighbor is at index 19523 with a distance of 12.0317
Scalable Product Quantization for Approximate Nearest Neighbor Search (SPANN)
It combines in-memory and disk storage, enabling the management of extremely large datasets while maintaining fast search capabilities. This hybrid approach is beneficial for systems that need to handle large-scale data efficiently.
SPANN benefits from batch processing architectures and the efficient use of GPUs and TPUs, as fast centroid searches in memory enhance efficiency and reduce wait times for disk read and write operations.
- Playground
Combines in-memory and disk-based storage. Centroids are kept in memory, while detailed data is stored on disk.
NOTE: SPANN is a complex system designed for industrial-scale applications and doesn’t have a direct Python implementation available. However, we can simulate the core concept using FAISS with Product Quantization (PQ).
import numpy as np
import time
from memory_profiler import memory_usage
import faiss
# Parameters
num_vectors = 100000
dimensionality = 128
nlist = 100 # Number of clusters
m = 8 # Number of subquantizers
# Generate and save dataset
np.random.seed(42)
dataset = np.float32(np.random.random((num_vectors, dimensionality)))
np.save('dataset.npy', dataset)
# Measure indexing time and memory usage
start_time = time.time()
index_memory_before = memory_usage()[0]
mapped_dataset = np.load('dataset.npy', mmap_mode='r')
quantizer = faiss.IndexFlatL2(dimensionality)
index = faiss.IndexIVFPQ(quantizer, dimensionality, nlist, m, 8)
# Train the index
train_sample = np.array(mapped_dataset[:10000]) # Use a subset for training
index.train(train_sample)
# Add data to the index in chunks
chunk_size = 10000
for i in range(0, num_vectors, chunk_size):
data_chunk = np.array(mapped_dataset[i:i+chunk_size])
index.add(data_chunk)
indexing_time = time.time() - start_time
index_memory_after = memory_usage()[0]
index_memory = index_memory_after - index_memory_before
# Query vector
query_vector = np.float32(np.random.random((1, dimensionality)))
index.nprobe = 10 # Number of clusters to search
# Measure query time and memory usage
start_time = time.time()
query_memory_before = memory_usage()[0]
D, I = index.search(query_vector, k=1)
query_time = time.time() - start_time
query_memory_after = memory_usage()[0]
query_memory = query_memory_after - query_memory_before
print(f"SPANN Simulation:")
print(f"Indexing Time: {indexing_time:.6f} seconds")
print(f"Indexing Memory Usage: {index_memory:.2f} MiB")
print(f"Query Time: {query_time:.6f} seconds")
print(f"Query Memory Usage: {query_memory:.2f} MiB")
print(f"The nearest neighbor is at index {I[0][0]} with approximate distance {D[0][0]:.4f}")
# SPANN Simulation:
# Indexing Time: 0.964418 seconds
# Indexing Memory Usage: 99.83 MiB
# Query Time: 0.106257 seconds
# Query Memory Usage: 0.08 MiB
# The nearest neighbor is at index 97986 with approximate distance 9.6613
Hierarchical Navigable Small World (HNSW) with performance tracing
HNSW constructs hierarchical graphs to enable efficient and accurate searches. It begins with broad searches at higher levels, progressively narrowing down to more precise searches at lower levels. This method provides a balance between speed and accuracy, making it highly effective for various applications.
The hierarchical nature of HNSW allows for efficient distribution of computational loads. GPUs and TPUs can process different levels of the graph in parallel, significantly reducing search times while handling large datasets.
- Playground
Builds a hierarchical graph where nodes are vectors, enabling efficient approximate nearest neighbor searches.
import numpy as np
import time
from memory_profiler import memory_usage
import hnswlib
# Parameters
num_elements = 10000
dimensionality = 128
# Generate dataset
np.random.seed(42)
data = np.float32(np.random.random((num_elements, dimensionality)))
# Measure indexing time and memory usage
start_time = time.time()
index_memory_before = memory_usage()[0]
p = hnswlib.Index(space='l2', dim=dimensionality)
p.init_index(max_elements=num_elements, ef_construction=200, M=16)
p.add_items(data)
indexing_time = time.time() - start_time
index_memory_after = memory_usage()[0]
index_memory = index_memory_after - index_memory_before
# Query vector
query_vector = np.float32(np.random.random(dimensionality))
p.set_ef(50)
# Measure query time and memory usage
start_time = time.time()
query_memory_before = memory_usage()[0]
labels, distances = p.knn_query(query_vector, k=1)
query_time = time.time() - start_time
query_memory_after = memory_usage()[0]
query_memory = query_memory_after - query_memory_before
print(f"HNSW Indexing:")
print(f"Indexing Time: {indexing_time:.6f} seconds")
print(f"Indexing Memory Usage: {index_memory:.2f} MiB")
print(f"Query Time: {query_time:.6f} seconds")
print(f"Query Memory Usage: {query_memory:.2f} MiB")
print(f"The nearest neighbor is at index {labels[0][0]} with a distance of {distances[0][0]:.4f}")
# HNSW Indexing:
# Indexing Time: 1.124930 seconds
# Indexing Memory Usage: 11.95 MiB
# Query Time: 0.107202 seconds
# Query Memory Usage: 0.00 MiB
# The nearest neighbor is at index 8769 with a distance of 13.3470
Conclusions
The choice of vector indexing technique depends on specific application requirements, such as indexing speed, memory constraints, and query performance.
- IVF Indexing offers the fastest query times with moderate memory usage, making it a solid choice for applications prioritizing query efficiency.
- HNSW Indexing provides low memory consumption and competitive performance, suitable for memory-constrained environments.
- SPANN Simulation balances indexing time and query performance but requires more memory during indexing.
- Disk-Based ANN Simulation is excellent for rapid indexing and handling large datasets but at the cost of higher memory usage.
- LSH Indexing may need optimization or reconsideration due to its high indexing time and memory usage anomalies.
Careful evaluation of the performance metrics in the context of the application’s needs will guide the selection of the most appropriate indexing technique.